Your comments
The additional constrain only removes entries with null values, when
something went wrong during the observation. We are fixing the behavior,
so in the next days, also the initial query will work.
You are not loosing results since the GIRAFFE spectrograph used in
the MEDUSA multi-object spectroscopy mode, allows the observation of up
to 130 distinct targets simultaneously.
Hence, each science raw file generates N products, where N corresponds to the number of science fibres. N can be as high as 130.
Also,
for those observation blocks where more than one exposure exists, the
individual 1D spectra are stacked and also available as products.
For more information please check the products documentation: https://www.eso.org/rm/api/v1/public/releaseDescriptions/73
and the instrument pages and manuals available through https://www.eso.org/sci/facilities/paranal/instruments/flames/inst/Giraffe.html
Hi,
to avoid the error, please add the following constrain:
AND dec BETWEEN -90 and 90
in this way you get 7 files as result.
The reason is that the raw database is not as curate as the products one, we are going to look into this, thanks for reporting!
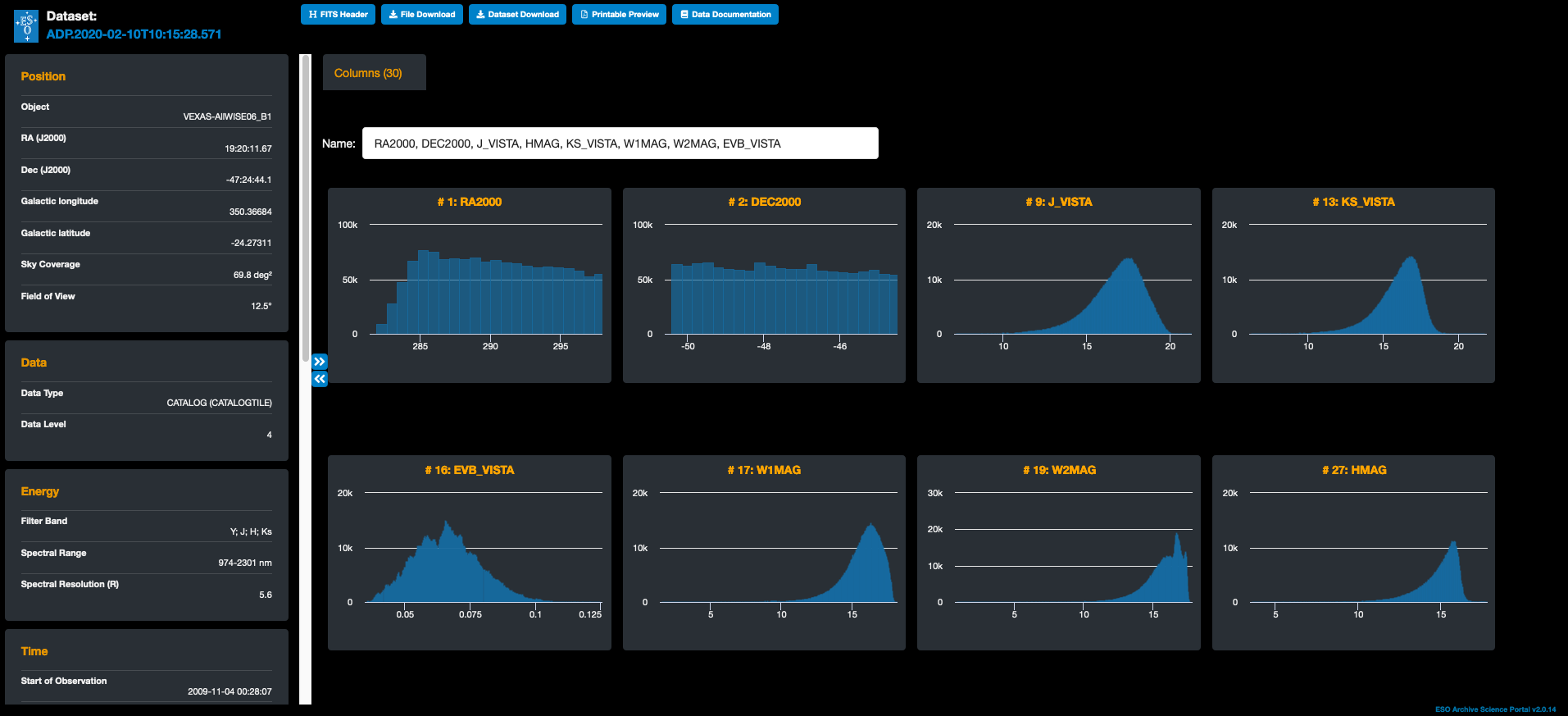
The walk-through on how to produce the Figure above using the programmatic access to the catalogue is illustrated in this forum entry (it requires the installation of the un-official ESOAsg python package).
It is also possible to explore the catalogue via the Archive Science Portal. This interface allows users to visualize the distributions of different columns in a catalogue tile in a comprehensive page, by clicking on the action Dataset details. An example of this is shown in the image above.
The TAP query to obtain all the XSHOOTER observations of the quasars part of the MAGG sample is:
SELECT Xshoot.dp_id
FROM
(SELECT * FROM ivoa.ObsCore
WHERE obs_collection = 'XSHOOTER') Xshoot,
(SELECT * FROM ivoa.ObsCore
WHERE obs_collection = '197.A-0384') MyColl
WHERE CONTAINS( Xshoot.s_region , MyColl.s_region)=1
Similarly, for the UVES spectra:
SELECT Uves.dp_id
FROM
(SELECT * FROM ivoa.ObsCore
WHERE obs_collection = 'UVES') Uves,
(SELECT * FROM ivoa.ObsCore
WHERE obs_collection = '197.A-0384') MyColl
WHERE CONTAINS( Uves.s_region , MyColl.s_region)=1
Customer support service by UserEcho